Update 2024-02-26: due to a serious typo all the results in this post were off by a factor of 100. Sequencing from individuals still looks promising, but by less than it did before. I've updated the numbers in the post, and added notes above the charts to explain how they're wrong. Thanks to Simon Grimm for catching my mistake.
While this is about an area I work in, I'm speaking for myself and not my organization.
At the Nucleic Acid Observatory we've been mostly looking at metagenomic sequencing of wastewater as a way to identify a potential future 'stealth' pandemic, but this isn't because wastewater is the ideal sample type: sewage is actually really—the joke writes itself. Among other things it's inconsistent, microbially diverse, and the nucleic acids are have degraded a lot. Instead the key advantage of wastewater is how practical it is to get wide coverage. Swabbing the noses and/or throats of everyone in Greater Boston a few times a week would be nuts, but sampling the wastewater gets you 3M people in a single sample. [1]
Imagine, though, that people were enthusiastic about taking samples and giving them to you to sequence. How much better would that be? What does "better" even mean? In Grimm et al. (2023) we operationalized "better" as RAi(1%): what fraction of shotgun metagenomic sequencing reads might come from the pathogen of interest when 1% of people have been infected in the last week. For example, in our re-analysis of Rothman et al. (2021) we found a RAi(1%) of ~1e-7 for SARS-CoV-2, which means we'd estimate that in a week where 1% of people contracted Covid-19, one in 10M sequencing reads would come from the virus. Let's say you were able to get throat swabs instead; what might RAi(1%) look like?
The ideal way to determine this would be to swab a lot of people, run untargeted sequencing, analyze the data, and link it to trustworthy data on how many people were sick. But while public health researchers do broad surveillance, collecting and testing swabs for specific things, as far as I can tell no one has combined this with metagenomic sequencing. [2] Instead, we can get a rough estimate from looking at studies that just sequenced sick people.
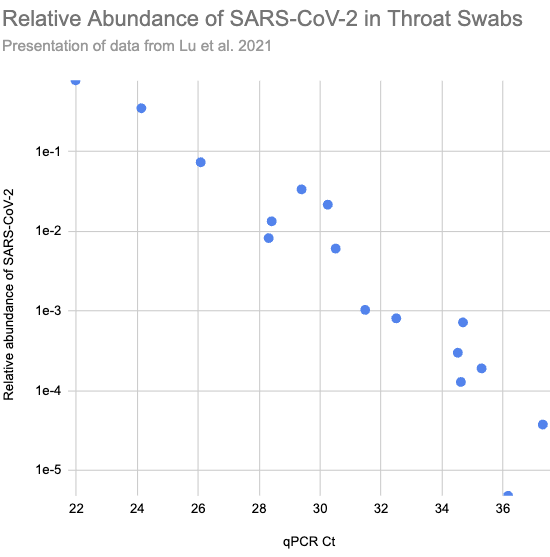
In Lu et al. (2021) they ran shotgun metagenomic RNA sequencing on throat swabs from sixteen Covid patients in Wuhan. Patients with higher viral load, and so lower Ct values on their qPCR tests (roughly the number of times you need to double the amount of SARS-CoV-2 in the sample until it's detectable), consistently had higher relative abundance:
Larger version of Fig 2.a.1, reconstructed from Supplementary Table S1)
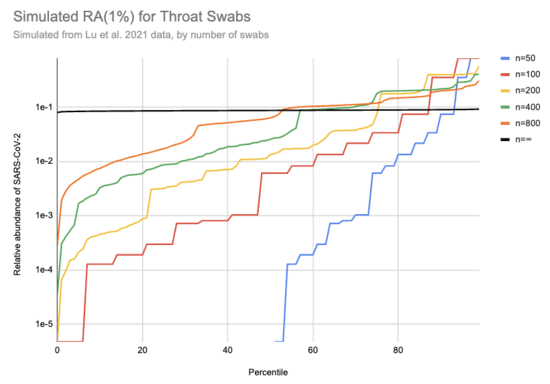
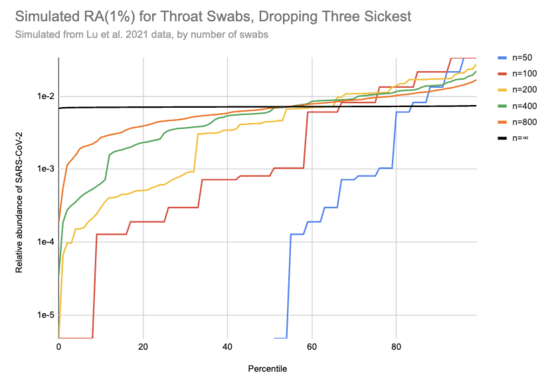
Imagine we got swabs from a lot of people, of which 1% were sick. What sort of relative abundances might we get? If we collect only a few swabs it depends a lot on whether we get anyone who's sick, and if we do get a sick person then it matters a lot how high their viral load is. On the other hand, if we collect a very large number of swabs then we'll just get the average across the population. Assuming for the moment that we can model "sick" as "similar to one of those sixteen hospitalized patients", here's a bit of simulating (code):
[EDIT: the y-axis on this chart is 100x too high. For example, the black line should be just below 1e-3]
This is 10k simulations, ordered by the relative abundance each gave. For example, if 1% of people are sick and you only swab 50 people then in half the simulations no one in the sample is sick and the relative abundance is 0, which is why the blue n=50 line only shows up for percentiles 50% and above. On the other hand, if we collect a huge number of swabs we end up with pretty consistently 0.08% of sequencing reads coming from SARS-CoV-2. With 200 swabs the median RAi(1%) value is 0.01%.
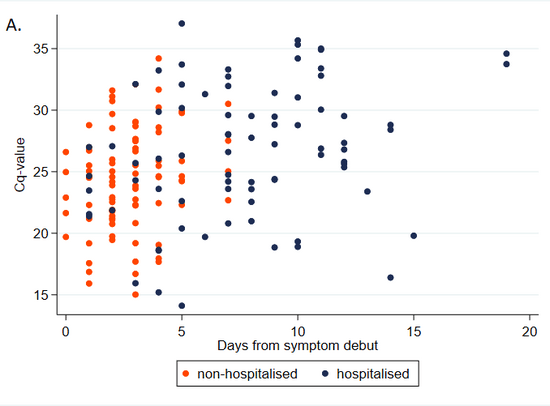
One major issue with this approach is that the data was collected from hospitalized patients only. Having a high viral load seems like the sort of thing that should make you more likely to be hospitalized, so that should bias Ct values down. On the other hand, people tend to have lower viral loads later in their infections, and hospitalization takes a while, which would bias Ct values up. Here's a chart illustrating this from Knudtzen et al. (2021):
Note that Cq and Ct are different abbreviations for same thing.Is there a paper that tells us what sort of Ct values we should expect if we sample a broad swath of infected people?
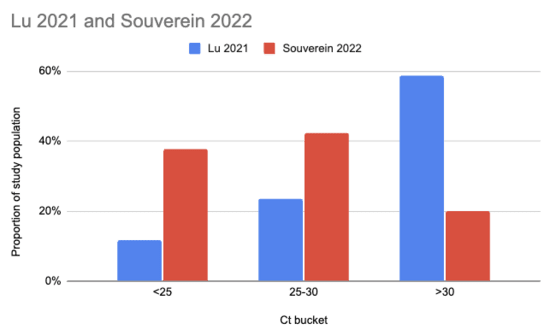
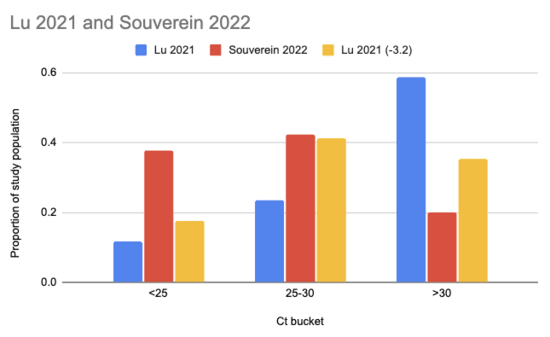
Souverein et al. (2022) looked at a year's worth of SARS-CoV-2 PCR tests from a public health facility in the Netherlands. The good news is these tests averaged two days from symptom onset and they got results from 20,207 people. The bad news is we only have data from people who decided to get tested, which still excludes asymptomatics, and these were combined nasopharyngeal (NP, "deep nose") and oropharyngeal (OP, "throat") swabs instead of just throat swabs. Still, pretty good! Comparing their Ct values to what we see in Lu et al. (2021), it looks like viral loads are generally a lot higher:
There are two issues with taking this chart literally. One is that the combined swabs in Souverein should generally have given lower Ct scores for the same viral load than throat-only swabs would have given. A quick scan gives me Berenger et al. (2020) where they found a median Ct 3.2 points lower for nasopharyngeal than throat samples, so we could try to adjust for this by assuming the Lu Ct values would have been 3.2 points lower:
The other issue, however, is worse: even though it's common to talk about Ct scores as if they're an absolute measurement of viral load, they're dependent on your testing setup. A sample that would read Ct 25 with the approach taken in one study might read Ct 30 with the approach in another. Comparisons based on Ct within a study don't have this problem, but ones across studies do.
So, what can we do? My best guess currently is that the Lu data gives maybe slightly lower relative abundances than you'd get sampling random people, but it's hard to say. I'm going to be a bit unprincipled here, and stick with the Lu data but drop the 20% of samples with the highest viral loads (3 of 16) to get a conservative estimate of how high a relative abundance we might see with throat swabs. This cuts RAi(1%) by a factor of ten:
[EDIT: the y-axis on this chart is 100x too high. For example, the black line should be just below 1e-4]
I really don't know if this is enough to where the remaining samples are a good representation of what you'd see with random people in the community, including asymptomatics, but let's go ahead with it. Then with 200 swabs the median RAi(1%) value is now 4e-5, a ~400x higher relative abundance than we see with wastewater. [3] If you could cost-effectively swab a large and diverse group of people, this would allow surveillance with much lower sequencing costs than wastewater. But that's a big "if": swabbing cost goes up in proportion to the number of people, and it's hard to avoid drawing from a correlated subgroup.
Thanks to Simon Grimm for conversations leading to this post and for sending me Lu et al. (2021), to Will Bradshaw for feedback on the draft and pointing me to Knudtzen et al. (2021) and Souverein et al. (2022), and to Mike McLaren for feedback on the draft.
[1] Technically it gets you that in two samples, since Biobot tracks
the North System and South System separately. But you can combine them
if you want simpler logistics.
[2] If you know of someone who has, or who would if they had the money for sequencing, please let me know!
[3] Pathogen identification would also be much easier with swabs, since it's a far simpler microbiome and the nucleic acids should be in much better condition.
Comment via: facebook, facebook, lesswrong, the EA Forum, mastodon

Thank you for this write-up, very interesting. I'm excited to see more investigations of different surveillance systems' potential.
Hopefully, the SIREN 2.0 study, running this winter, will generate some more data to answer this question.
A few questions now I've had time to consider this post a bit more. Apologies, if these are very basic, I'm pretty unfamiliar with metagenomics.
First, how do you relate relative abundance to detection probability? I would have thought the total number of reads of the pathogen of interest also matters. That is, if you tested the entire population you would have some reads on every pathogen even if the relative abundance of some pathogens is very low.
Relatedly, does the cost of the sequencing scale roughly linearly with the relative abundance required? That is, if your 40,000x figure is correct, would that imply swabbing is ~40,000x cheaper than wastewater?
Finally, could you please expand on your figure of percentiles vs relative abundance? Why does the number of swabs affect the relative abundance? If you double the number of swabs, I would expect that both the total reads and the number of SARS-COV-2 reads double, hence holding the relative abundance constant. Perhaps it's that the variance in the number of individuals infected and their viral load increases but the mean of all the lines is the same, is that correct?
And now one point of disagreement.
I don't think this is that big an "if".
1% incidence per week is extremely high. For context, SARS-CoV-2 at 1% incidence per week implies a prevalence of 2-3%. The UK, which had a middle-of-the-road rich world pandemic, peaked at this prevalence just before implementing a lockdown in the January 2021 (Alpha variant) wave.
The main scenario discussed (in the context of NAO) is stealth respiratory pathogens. Respiratory pathogens spread fairly indiscriminately because they spread in public areas. This is unlike pathogens spread through close contact (eg: HIV or mpox), which can be contained in smaller communities more easily.
In other words, a respiratory pathogen at 1% incidence per week is as widespread as SARS-CoV-2 at its worst in the pre-vaccine era, and spreading roughly as indiscriminately (it could be spreading slower). I don't see it being contained in hard-to-target populations.
Furthermore, wastewater sequencing doesn't fully solve this. Unless you cover all wastewater across the globe, you're still not covering everything. If you do large cities it's probably not as bad as individual swabbing but I'm not sure it's much better. Other settings, such as airports/airplanes, seem at least as bad as the populations you might target for convenient swabbing (eg: hospital or primary care patients).
So while the sample might be a few times either way biased, I'm sceptical this is bridging a 40,000x gap (maybe 40,000x isn't the relevant benchmark here - see comments previously).
Lots of great questions!
Thanks for pointing this out; I hadn't seen it and it's super relevant. I don't see what sample type they're using in the press release, but any kind of ongoing metagenomics to look at respiratory viruses is great!
It depends on your detection method, but modeling it as needing some number of cumulative reads hitting the pathogen is a good first approximation.
If you think it would take N reads of the pathogen to flag it then if you know RA(1%) and the exponential growth rate you can make a back of the envelope estimate of how much sequencing you'd need on an ongoing basis to flag it before X% of people had ever been sick. For example, if you need 100 reads to flag, it doubles weekly, and RAi(1%) is 1e-7 then to flag at a cumulative incidence of 1% (and current weekly incidence of 0.5%) you'd need 100/1e-7 = 1e9 reads a week.
(I chose 1% cumulative incidence and weekly doubling to make the mental math easier. At 1% CI half the people got sick this week and half in previous weeks, and the cumulative infection rate across all past sequencing should sum to 1%, so we can use RAi(1%) directly. Though I might have messed this up since I'm doing it in my head lying in bed.)
If you collected a large enough sample volume and sequenced deeply though, yes.
It doesn't, for three reasons:
Sequencing in bulk is a lot cheaper per read. You might pay $13k for 10B read pairs, or $1k for 100M. But that's just ~10x.
Some components (lab time, kits) vary in proportion to the number of samples and don't go up much as your samples are bigger.
It's only your sequencing costs that vary with relative abundance, and while with wastewater I expect the cost of sequencing to dominate that's not the case for any other sample type I can think of (maybe air?) If you're sampling from individuals the cost of getting the samples is likely quite high (we were recently quoted $80/person from a contractor, and while I think we can do better if you want 1k people per pooled sample it's almost certainly more expensive than the sequencing charge).
Some people have vastly higher viral loads than others, and the relative abundance you see for a pool depends on whether you get some of these people. Your intuition would be correct for pools large enough that this variation was no longer relevant.
Sorry, I was unclear! The easiest way to collect a pooled sample is the walk around some building and sample everyone. This gets you a big sample pretty cheaply, but it's not a great one if you want to understand the containing city because it's likely that many people in the building will get sick on a similar timeframe. The sample members are too correlated in their exposure.
In your UK example, I'm guessing you could sample some office buildings of 1k people and find 0 cases and others and find 200 cases.
To avoid this you need broader sample collection, but that's logistically more difficult and so more expensive.
Airport arrivals would be great, though that's a difficult setting to work in.
I'm also skeptical! I think sampling from individuals is extremely promising. It seems like you ought to be able to get down to more like $2/person in which case a pool of 1k costs you $2k in collection. Then add in $1k for sequencing and you're still well above wastewater. But my initial attempts to partner with people already doing sampling haven't turned up good leads.
Thank you for that very detailed reply Jeff, I learnt a lot about how to think about costing this.
I agree this is true for an office block, but I would think you can do much better without much cost. For example, if you use a high-traffic commuter train station or supermarket I would guess you get a fairly broad cross-section of the city. They'd be somewhat uncorrelated (different home locations with children at different schools, different offices etc.) although obviously the geographical component is still there. Perhaps similar to wastewater though? You could do multiple locations as well though.
These numbers are maybe optimistic, but not ridiculously so.
The Coronavirus Infection Survey (big UK study which I've worked on) cost ~£1b for ~11.5m swabs (Excel sheets with data from Mar 2023 and historical data). Works out as ~$100 / swab.
Very likey overestimated upper bound though because that is a proper random sample of the whole population, with ~9.5m of the swabs collected by study workers going to houses. I think this budget might exclude the cost of PCR testing (done individually, not pooled) and a lot of time spent running / analysing the data.
Definitely! Right after writing to you I started thinking about this, estimating costs, and talking to coworkers; sorry for not posting back! I do think something along these lines could work well.
My main update since then is that if you do it at a transit station you probably need to compensate people, but also that a small amount of compensation doesn't sink this. Giving people $5 or a candy bar for a swab is possible, and if a team of two people at a busy transit station can get 50-200 swabs in an hour that's your biggest sample acquisition cost. I still think $1k is practical for the sequencing.
I'm trying to come up with examples of people doing something similar, which we'd want for presenting this to the IRB. Two examples so far:
XpresCheck for COVID tracking at airports (site, [consent brochure] (https://www.xprescheck.com/xpresresources/CDC_COVID_Testing_Brochure.pdf))
Various companies that sample for bone marrow compatibility testing (ex: Be The Match)
Do you know of anything else that feels similar to this? People in public areas collecting biological samples from volunteers (perhaps lightly compensated).
Afraid not. The closest I can think of is collecting samples from healthy volunteers without any benefit to them, but not in public areas. In particular, I'm thinking of swabbing in primary health settings (eg RGCP/UKHSA run something like this in England, I can't remember if it only includes those with respiratory symptoms) and testing blood donations (normally serological testing looking for antibodies). REACT (run by Imperial College) did swabbing for COVID via postal recruitment.
A bit of an aside, so maybe not of interest, however, this made me think of serological testing of residual blood samples. That is, when blood is collected for testing (for any clinical reason), not all of it is used in the tests, and the remaining (residual) part is tested. Here, there are no sample collection costs (the blood was collected anyway). However, it doesn't map exactly because you don't swab people without respiratory suspicion but you might take blood (eg anemia). Maybe there is an opportunity for either testing blood samples for pathogens (but I have no idea what that looks like) or samples taken for other respiratory reasons (but then you need to think about co-infection, ie does infection with influenza make you less likely to have another respiratory infection).
Finally, some shameless self-promotion. I'm currently nearing PhD competition with nothing lined up. If there are projects looking at these sorts of questions interested in modelling / stats / epidemiology input I'd be very interested, please DM. Please ignore this if unappreciated.
The original version of this post had results from a simulation where the key results were off by a factor of 100. See the update at the top of the post for more.
Feel free to message me if you're interested in going deeper into what a typical viral load might look like. I can generate trajectories, based on the data from the ATACCC study. Note that this is in viral RNA copies, not Ct values - they did the conversion as part of that study.
Thanks! I'm most interested in viral load in the sense of the relative abundance you get with untargeted shotgun sequencing (since you need sequencing (or something similarly general) to detect novel threats and/or avoid having a trivially-bypassable detection system) but there's not much literature on this.